Bioinformatics:
Converting Data to Knowledge
Gio Wiederhold
Stanford University
This note documents introductory remarks presented at a workshop sponsored by the National Research Council, 16 February 2000, in Washington DC.
We are competing this week with the announcement of Windows 2000 and the excitement engendered by the Internet. But the topics discussed in our meeting will be of lasting value, and not be made obsolete by a new release next year.
Data, Knowledge and Information
Bioinformatics has become the accepted term for a new field in science, covering research, development and applications of information science and technology to biology and medicine. Information is created at the confluence of knowledge and data. Data is obtained from observations, and its values should be objectively verifiable. Knowledge is gained through processing of observations, gained by experience, teaching, and the more formal processes we will focus on. Knowledge is compact, it applies to many data instances.

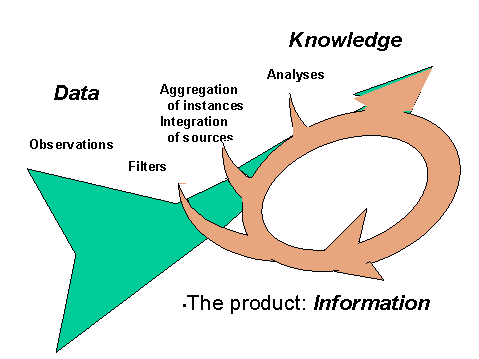
Figure 1: Interaction of Data and Knowledge
Creating knowledge from data is not a one-way path. The processes that convert data to knowledge themselves require knowledge (Figure 1). We need knowledge to select and filter appropriate data from an ever-increasing flow of observations, and assess their correctness. We need knowledge to classify instances and aggregate their parameters. We need knowledge to integrate the plethora of diverse sources. We need knowledge to select analyses and understand the meaning of the results. Knowledge is needed to understand and abstract the results into effective knowledge and information.
We define information, following Shannon, as having novelty, and affecting the state of the world. We want to change the world in many ways. Bioinformatics is generating information at Internet speed.
The information we gain allows us to learn about ourselves, about our origins, and about our place in the world. We have learned that we are quantitatively strongly related to other primates, mice, zebrafish, fruit flies, roundworms, and even yeast. The findings should induce some modesty, by learning seeing how much we share with all living organisms.
The information we are gaining is not just of philosophical interest, but also intended to help humanity to lead healthy lives. Knowledge about primitive organisms provides much information about shared metabolic features, and hints about diseases that affect humans in an economical and ethically acceptable manner.
Applying the knowledge can lead to new scientific methods, to new diagnostics and to new therapeutics.
Integration and heterogeneity
Knowledge from many scientific disciplines and their subfields has to be integrated to achieve the goals of bioinformatics. The contributing fields are autonomous. They set their standards, work at their level of abstraction and set their scope. Their heterogeneity inhibits integration, often because their terminologies differ.
The terms they use to communicate within the field will not match the terminology used be external collaborators. Terms used in field denote concepts in a in a field-specific granularity and its meaning is circumscribed to a field-specific scope. These terms change as fields grow and new knowledge is acquired.
We may wish to overcome the problems of heterogeneity by having standards. We depend greatly on standards for our infrastructure: computing hardware, communications, programming languages and operating systems – although Windows 2000 is bound to deliver some disappointments.
But standards require stability. Yesterday’s technological innovations have become today’s infrastructure. Progress in all aspects of bioinformatics will be rapid for some time, but sharing, integration, and aggregation of information throughout is essential and that means we have to deal explicitly with heterogeneity of our sources for information.
Making errors due to terminological problems is common. Effective recall on the world-wide-web is limited by a flood of irrelevant, obsolete, and even wrong information. The poor precision arises mainly because the terms used for searching are not constrained to their contexts. The large quantities of data that we deal with in bioinformatics require precision, since the cost of following up on every false-positive finding is relatively high, often greater than the process which generated 1000’s of findings in parallel.
Scaling bioinformatics computations
We have been dealing with the 4 billion base pairs of the human genome, and shorter, but significant strands from other species. As we move to diagnostics and therapeutics w must match abnormalities to about 10 000 protein-coding regions s and their disease-causing variations. Genetic variations effect every single one of our 6 billion world population.
At the same time, researchers in chemical genomics are generating an increasing fraction of the few million small organic molecules. These molecules may be used in pharmaceuticals to turn protein generation or protein effectiveness on and off, affecting the several hundred metabolic pathways that control our six billion lives, and many more in future generations (Figure 2).
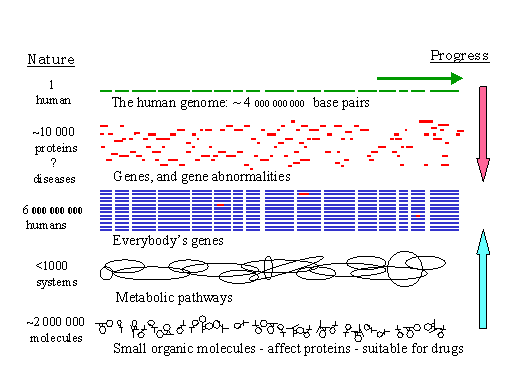
Figure 2. Quantities of some potential data objects in bioinformatics
We note that the volume and diversity of the biological and chemical material bioinformatics must deal with is immense, but finite. To make the linkages among them means solving many puzzles many times. Being selective and smart will help. Precision will be critical -- whenever we deal with 100 000’s of instances, even a 1% false positive rate means following up on 1000 false leads, easily overwhelming our research capabilities. When those leads involve people we must be especially careful.
Keeping knowledge about individuals private
Having knowledge carries responsibilities. Maintaining the privacy of patients and research subjects requires ethical insights, that may transcend rational scientific reasoning. How will people feel about your knowledge about them? When you know their genetic make-up you can draw inferences about their physical and psychological propensities. Although privacy is hard to formalize, but that does not mean it is not real to people. Perceptions count. Of course, there are also real dangers, as insurance scams, and effects of loss of privacy on personal relations, as release of pregnancy or STD tests.
Privacy concerns also intersect with the more complex issue of performing diagnostic tests for diseases that have no therapy available. In this instance there is also the issue of informing the patients and the patient’s family prior to diagnostic tests which could generate knowledge that might be problematical to handle. I won’t address this issue further here, but there are certainly no easy answers. Being sensitive to individual needs is an essential initial step.
To protect privacy security has to be provided. In science we have collaborative situations, and traditional technology developed for simple military and commercial settings is inadequate. Private data will not be secure unless the scientists who hold the data make its security an ongoing personal concern. The responsibility for privacy of data about people held by scientists cannot be simply delegated to specialists.
Simple solutions are based on good-guys versus bad-guys models, and intended to deny access to the bad, or any unknown requestor. However, in science we have collaborators, to whom we cannot simply deny access. But collaborators cannot be simply trusted with all information. When access is permitted we still must filter and log all information flowing out of our systems (Figure 3). An anonymized medical record, covering several dated visits has a unique signature that still can be used to identify the individual patient. Ancillary data, as a patients profession, can simplify unwanted identifications. Embarrassing data can easily be inadvertently released as part of an innocuous record collection. Misfiled data, an uncomfortably frequent occurrence in health care, also loses protection when solely controlled through access right management.
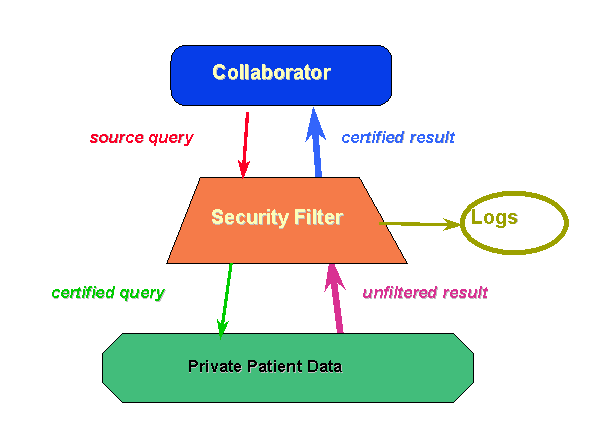
Figure 3. Filtering the release of patient data, as well as access.
We have many examples of the inadequacy of commercial systems, as when a music web site permits a customer to take out 250 000 credit card numbers instead of one MP3 encoded song. Simply checking what is being taken out could have prevented that disaster. We don’t want scientific progress in bioinformatics to be damaged by bad publicity because of lack of concern for patients’ privacy.
People
We must also be concerned about the people in our domain. The demand for researchers and staff in bioinformatics is high, exceeding available resources. More people need to be trained, and those in the field must be encouraged to stay, and enabled to be productive.
There is a lack of training opportunities – the shortage is both in programs and teachers, as well as in available and adequately prepared trainees.
Being in multi-disciplinary field is scary. Faculty worries about tenure when they bridge existing departments. Students may be required to satisfy fundamental requirements of multiple departments as well. Traditional engineering accreditation needs updating to allow graduates of engineering departments to specialize into bioinformatics relevant fields.
Combining departments requires resolving salary and growth differentials in biology and Computer Science. Bioinformatics must compete for bright students with World-Wide-Web visions. Some leading institutions are moving aggressively, and should become the models for a broader range of institutions.
Lets continue now with the central focus of our workshop: Bioinformatics:
Converting Data to Knowledge, with the means: People and the product: Information.
===================================================
Closing Observations
The workshop covered the generation and integration of biologic databases, their interoperation, and integrity maintenance. It addressed modeling and simulation, data mining, and visualization of the results. There was little emphasis on clinical effects. It does take a long time to close the data loop, where the results of bioinformatics research transition into medical practice. Only then we will be able to collect observations taken from patient populations, validating findings now inferred from concepts based on more abstract models. Initiating that loop is also extremely costly and requires a concentration of resources found only in the pharmaceutical industry. Improvements in management and perhaps some constraints on claiming ownership of intellectual property at pre-clinical stages might be beneficial for effective industry-academic research collaboration.
I observed two recurring topics in the workshop and during discussions in the breaks:
1. The importance of keeping knowledge about the data accessible, sharable, and subject to correction.
2. The importance, and the difficulties of exploiting data from multiple sources.
These observations are of course my own, determined by my background. Other participants are likely to have gone home with other conclusions.
Data and models
In bioinformatics the volume and diversity of data to be processed to reach conclusions is so massive that no single primary source is adequate. Primary data for each type of data is already collected at many sites, and collaboratively aggregated. These aggregations are integrated with other sources and data types to gain meaning and significance. Having a collaborative, many-faceted research structure means that processing to gain knowledge occurs on secondary, aggregated and derived data. Since that data is removed from its original context, there is a great need for documentation about the origins and subsequent transformation of the data, or its provenance.
Knowledge about data or metadata has many aspects and forms, and requires careful curation and maintenance. Anytime that data are made available, its entire provenance, from original observation, its filtering, transformations and aggregations, and any application of corrections by humans or automatic processing, must also be available. Both data and metadata must be available in electronic form; the volume of information being processed does not allow that metadata be relegated to a paper trail.
The provenance also implies models of how and why the data was being obtained and used initially. Such models can convey objectives, and understanding of the objectives for collection and transformation conveys an understanding of strength and weaknesses in a primary or derived data collection. Using the data for a different objective is likely to make the data less precise for that new context. For instance, GenBank and PDB collect data as a by product of scientific publication. Novelty and correctness is valued more than completeness in publications. This means that, for instance in PDB, some of the binding sites, that were observed and documented in the basic protein crystallography sources, are missing. Similarly, metadata used for bibliographic indexing, as NLM's MeSH, does not deal well with the explosion of knowledge in bioinformatics.
Processing of derived, secondary data is essential in bioinformatics, since for most analyses complete data cannot be obtained from a single primary source. Models, defining secondary processing, serve in selecting and classifying the data for a specific analytical objective. Curation of these secondary data may append or fix annotations, sometimes by going back to primary sources, sometimes by inference from other secondary sources, as needed for the specific research objective. These processes create intermediary databases of considerable added value, through the application of expertise focused on some objective.
Prof. Gelbart drew a simile for data collection, namely hunter-gatherers (collecting data on an ad-hoc basis) moving to an agronomic society (systematic mass production of data). That simile can be extended to medieval guilds -- the predecessors of our professional societies -- sitting around the market square, where the farmers deliver their goods, and specialists select, aggregate and combine goods, reselling them as wholesalers. Our collaborative research enterprise is reaching the stage that intermediaries are becoming essential.
In this research model of secondary processing ownership of data and metadata is shared. This concept is already recognized in the publication domain, where a collection can be assigned copyright, although all the individual source papers also retain their copyright protection.
Processing many instances to gain knowledge was shown not to be limited to text, at UCLA aggregation of images provides visual insights hard to put into words.
Integration and heterogeneity
The breadth of sources requires recurring integration of data to be a focus of knowledge generation. The diversity of the sources requires dealing with their heterogeneity at many levels, causing at least as much pain as peeling an onion.
The heterogeneity of access at the physical is being addressed by computer technologists and scientists. Differences in operating systems, database systems, and computer languages are becoming less of a barrier. The requirements of the Internet are reducing communication and interface barriers, since nearly everyone wants to be seen and heard on the web. Many databases can be remotely accessed through application programming interfaces (APIs), but simpler technology is replacing the programmed approach. Developments of HTML, which provided a common on-line publication interface for the web, are leading to XML, a common data representation interface. XML provides a means to attach metadata, lacking in APIs. XML metadata is expressed through tags for each data element, and the structure of the tags can be made available as a data type definition (DTD). Several efforts are underway to define DTDs for areas in bioinformatics, for instance in neurosciences.
However, each technological improvement only moves the bottleneck to a higher level stage. XML provides a consistent tagging format for metadata, but the actual meaning of the tags still depends on their contexts, and cannot be centrally specified for all areas in bioinformatics. I heard the need for an XML debabelizer being expressed. In practice, we have to learn to communicate among areas in bioinformatics without demanding global standards or single solution. Technically this means that we will establish articulations, as needed, in the intersections where knowledge can be produced from bioinformatics data, rather than a globally consistent federation.
================================================